CpG islands are generally known to be unmethylated. But there are so many exceptions in the measured data you can easily find. Let's examine the DNA unmethylation with GSE97484, which is a multi-omics data set composed of subsets of DNA methylation and ChIP-Seq of H3K4me3 or H3K27ac.
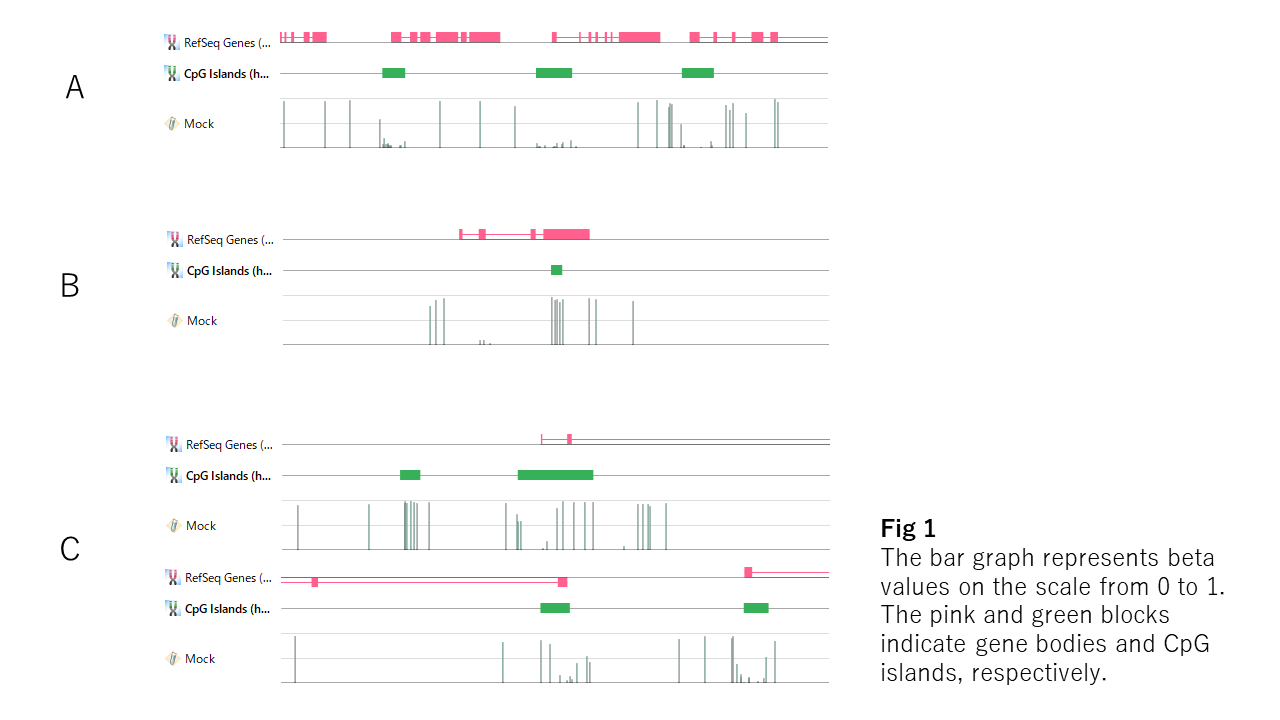
If you see the beta values on a genome browser, you see many cases like Fig1-A. The unmethylated regions look like bottoms of valleys, and the edges are steep and tightly linked with the boundary of the CpG island. But you also find exceptions. The entire region of the CpG islands is sometimes methylated (Fig1-B), or the border of the unmethylated region doesn't match with that of the CpG island (Fig-C). You also find unmethylated regions outside the CpG island (Fig1-B). So, CpG island doesn't seem to be essential for the DNA unmethylation.
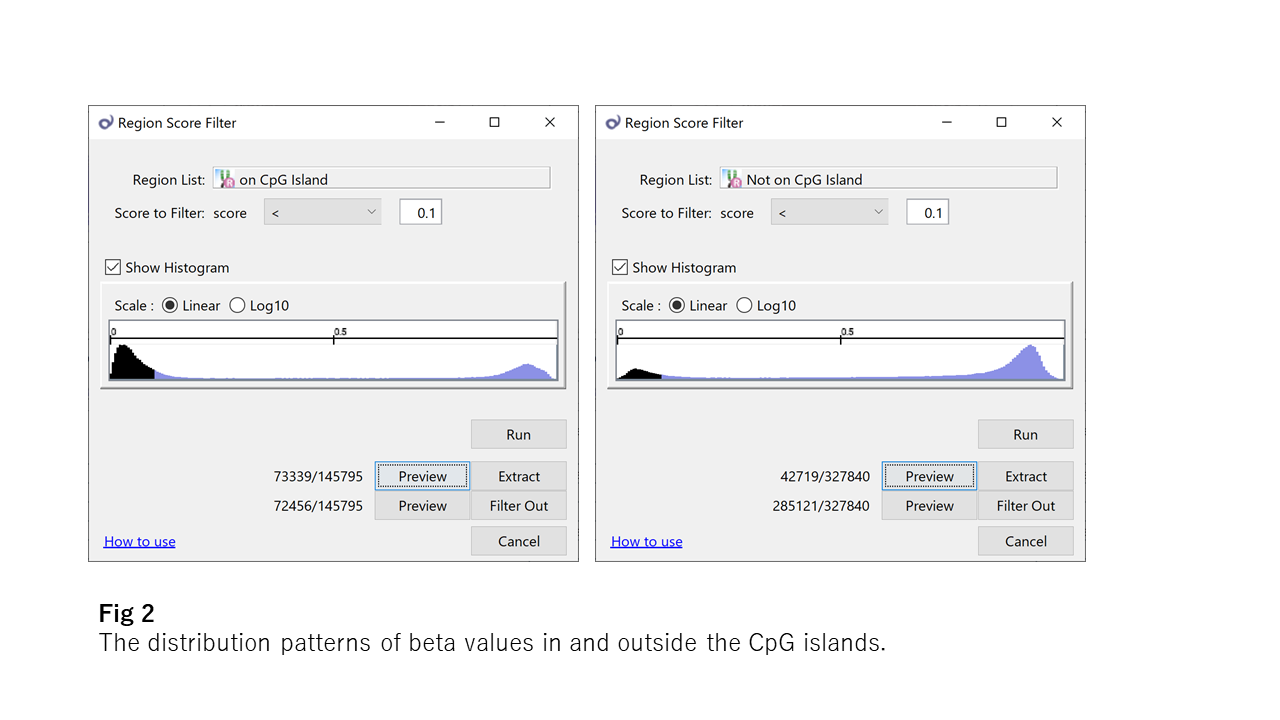
If you look at Fig2, you understand how unmethylated the CpG islands are. Half of them are <0.1, and it's in contrast to outside CpG islands. But you also understand why there are many exceptions because there is a small peak on the opposite side.
Now take a look at Fig3, which shows the distribution patterns of beta values in and outside H3K4me3 ChIP-Seq peaks. H3K4me3 (the tri-methylation at the 4th lysine residue of the histone H3 protein) is a famous epigenetic modification. The H3K4me3 area looks more strictly unmethylated than the CpG island.
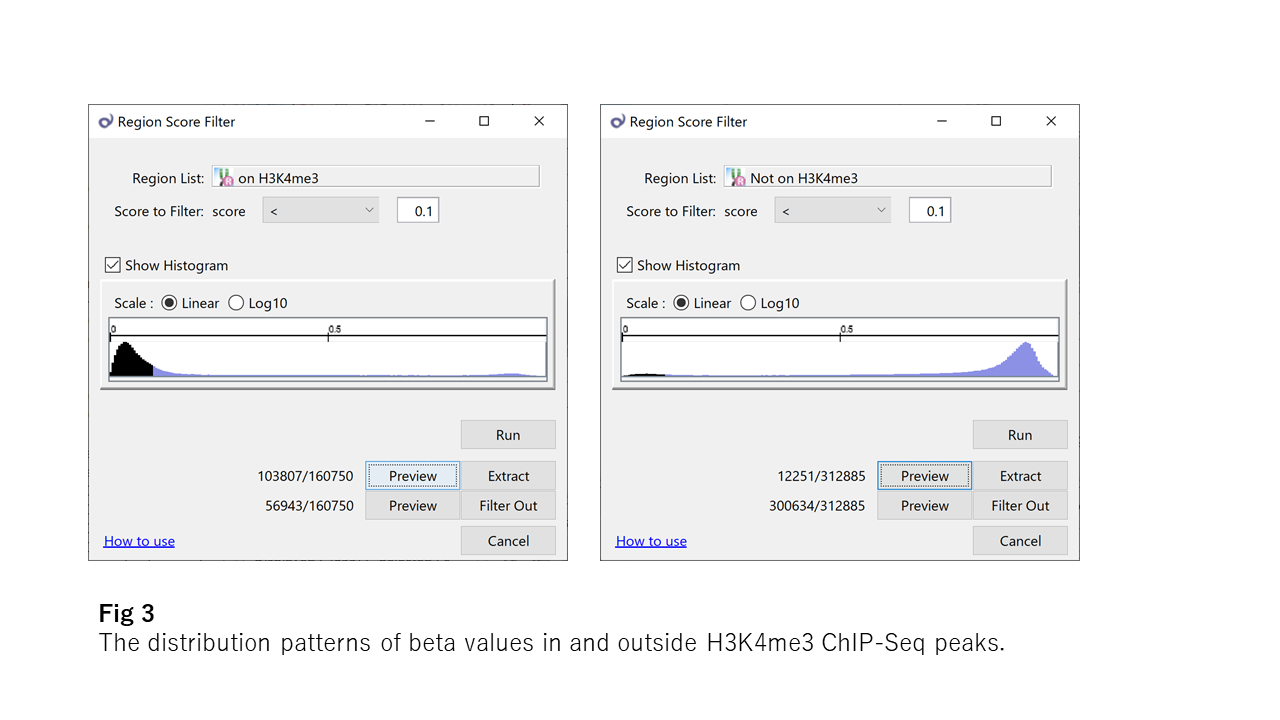
Fig4 shows the beta value distributions of overlapping or not-overlapping sites of CpG islands and H3K4me3 ChIP-Seq peaks, respectively. H3K4me3 looks to be essential for the unmethylation of CpG islands because, even in CpG islands, sites outside H3K4me3 peaks are mostly methylated. And H3K4me3 strongly affects the DNA unmethylation even outside the CpG islands.
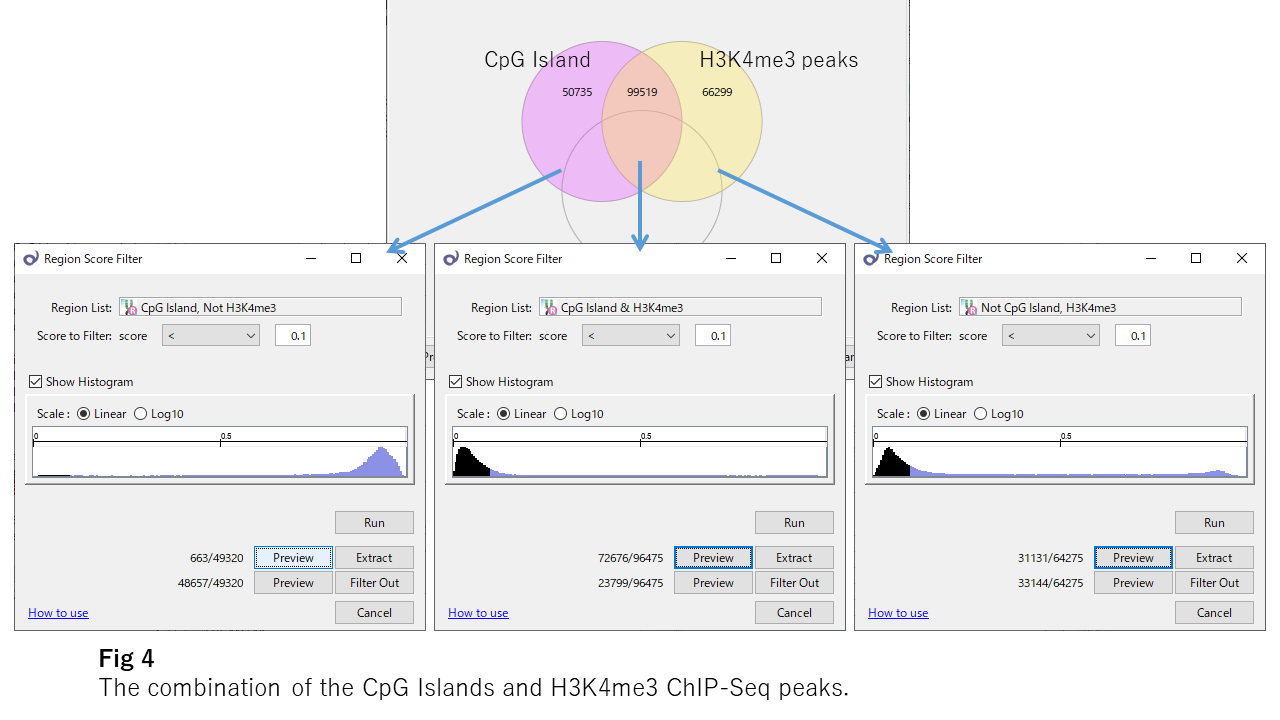
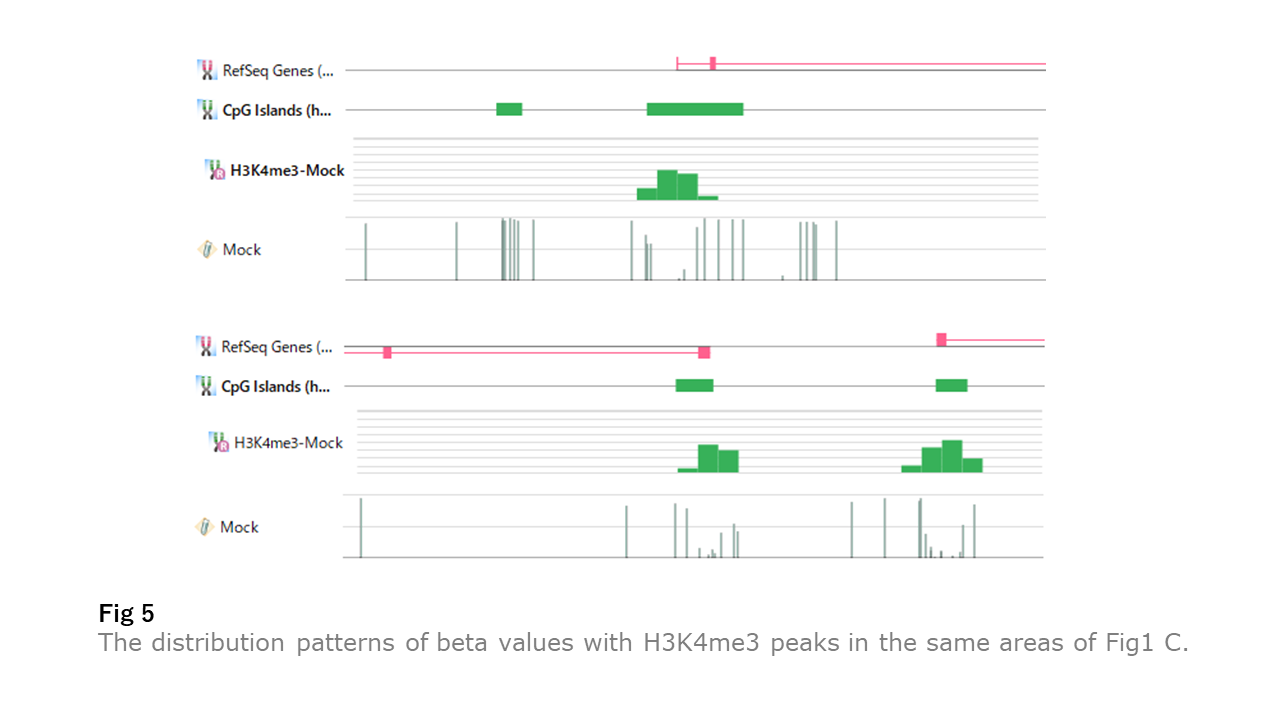
Fig5 illustrates the same areas of Fig1 C with H3K4me3 peaks. The unmethylated regions correspond more with H3K4me3 peaks than the CpG islands. Besides, we saw the apparent link between the unmethylated regions and H3K4me3 peaks in Fig1 B cases.
Information:
The ChIP-Seq data of GSE97484 is provided in bigwig format files. Please learn how to import bigwig files to Subio Platform.