
University College Cork, Lab Researcher
Dr. John Mac Sharry
It is an excellent user interface and the tutorials are brilliant.
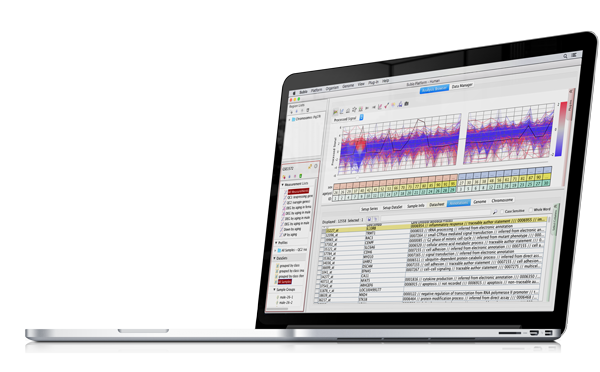
Subio Platform is a data analysis software for RNA-Seq, microarray data. And it works fine for the integrative analysis of multi-omics data sets regardless of the type or manufacturer of the measurement system.
What to learn;
Command Operation?
Or Data Analysis?
To download Subio Platform, enter your information below, agree to the terms and conditions, and click the download button.
As you know, R (Bioconductor) and Python are powerful statistical tools widely used in this field. But researchers also need a set of various utilities to challenge the complexity of life science. Subio Platform offers the whole without a pricy license fee like GeneSpring. Plug-ins are not free but reasonable compared to free bioinformatics tools because you must pay the high implicit learning- and maintenance-cost. Moreover, you might be surprised that you can accomplish almost all analyses without plug-ins for preliminary analysis.
We provide lots of free information about usage, tutorials, troubleshootings, and case studies from this website so that you can learn by yourself. We also offer charged services of online training and outsourcing data analysis services.
Now please let us introduce you Subio Platform software.
If you drag on a chart, you'll instantly know what those genes are. Likewise, if you reversely search for gene names or keywords, you'll immediately see how those genes behave on the graph. In such ways, it is much easier to understand the data than to operate with commands. And all views are associated so that you can grab the data from multi-angles.
The omics data is so complex that you must fully use such sophisticated graphical aids because understanding data is fundamental. Unfortunately, many training programs about omics data analysis tend to underestimate viewing data while focusing too much on statistical commanding. However, you have to understand the data adequately first of all.
| Scatter Plot (Measurement) View |
Detail
<p>Scatter Plot (Measurements) View interactively visualizes data of arbitrary two of sample groups. Drag a sample group from the series panel, and drop upon vertical or horizontal axis to set. You can compare how similar or different between signals between the two. Or you can see the relationship between signal intensities and log ratios of one sample group.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use it.</p>
|
|---|---|
| Line Graph View |
Detail
<p>Line Graph View is useful to visualizes changes of signals among Sample Groups. It's interactive and you can select genes just by drag and drop on the chart. The selected genes are emphasized in the Annotations table in the lower panel as well. You can make a Measurement List of the selected genes by clicking on "Save as Measurement List" button, which is next to the camera icon.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use it.</p>
|
| Tree View |
Detail
<p>Tree View is for browsing a heatmap as a result of hierarchical clustering. A heatmap image on PDF is dead, but this view is interactive. You can traverse nodes, select genes under a node and select samples.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use it.</p>
|
| Pathway View |
Detail
<p>Pathway View overlays expression pattern as heatmaps or bar graphs on pathway images. You can drag on it to select genes or filter genes by selecting a measurement list. It helps biological interpretation.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use it.</p>
|
| Genome View |
Detail
<p>Genome View is a genome browser which is essential to analyze data related to genomic locations, like ChIP-Seq, Methyl-Seq, tiling array, CGH array, ChIP-chip, methylation array and so on. Even if you analyze gene expression data, you can use this View to see if the up- or down- regulated genes are located closely or not. If you see dense area of such genes, it may indicates they were controlled by changes of chromosomal structure or epigenetic status.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use it.</p>
|
| Scatter Plot (Samples) View |
Detail
<p>Scatter Plot (Samples) View is for browsing PCA results, or samples according to your imported profiling. Dots represent sample groups composed of the selected DataSet. Notice that selecting another DataSet changes plots on the chart. You can drag to select sample groups, and Samples involved in the selected Sample Groups are also superimposed in Sample Info tab in the lower panel, and Tree View. These views interact each other.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use this.</p>
|
| Venn Diagram |
Detail
<p>You can combine measurement lists with Venn Diagram too, to extract genes at intersection, union or difference. Drag&drop a measurement list from Series panel to one of circle of Venn Diagram. If you'd like to combine more than 3 lists, open "# Overlapping" tab. You can input many measurement lists to combine.</p>
<p>Please request a free <a href="/support" title="/support">Online Support</a>, if you don't know how to use this tool.</p>
|
Undoubtedly, it would be a great help if you have a database of your own experimental data with massive omics data sets available from the web like GEO (Gene Expression Omnibus) and TCGA (The Cancer Genome Atlas). Indeed, nobody can thoroughly extract biological knowledge from omics data. So the legacy data sets are still juicy.
However, handling data sets in various formats generated by multiple systems might take time, and of course, it requires experience and skills. If it's hard, we can offer Data Analysis Service.
| Importing microarray data sets from GEO | Detail |
|---|---|
| Importing TCGA RNA-Seq data sets | Detail |
| Importing TCGA miRNA-Seq data sets | Detail |
| Importing TCGA DNA methylation array data sets | Detail |
If you are still looking for the file type you want to analyze, or if you face errors in importing files, please find solutions on the web or consider charged online training.

Human beings have yet to learn how to interpret omics data. So we absolutely need much more discussion involving biologists, not only bioinformaticians. However, it takes work for them to participate due to the unfriendliness of bioinformatics tools.
If you export the SSA file from Subio Platform, you can hand the raw data with final and intermediate results in a single file. Your collaborators rebuild the analysis environment only by importing the SSA file, and they can instantly start examining analysis results and re-analyzing from their perspectives. It enables deeper discussion and collaboration than PDF- and Excel-based handovers.

Although open-source and free software have supported bioinformatics, the problems of such tools also become apparent. For example, operating systems and middleware must keep updated to counter various threats. On the other hand, bioinformaticians abandoned their tools without maintenance, and it caused problems that researchers couldn't use the same software in their long-running studies. This problem happened because bioinformaticians' evaluations are biased toward developing new tools, and there is little incentive to maintain existing tools.
To solve this problem, we established Subio as an autonomously sustainable operating entity that receives a small license fee from plug-in users. Since the advent of microarrays, various measurement devices and analysis software have appeared and disappeared, but Subio has kept running without wavering from its original purpose. As a result, it becomes the only stable software platform to accumulate and integrate new and legacy omics data for analysis.
Subio also proposes to bioinformaticians an alternative distribution of their tools to keep them maintained.

Subio Platform offers valuable utilities and interactive GUI that R or Python don't provide, but conversely, it doesn't offer advanced statistical features. However, you can easily output data from Subio Platform and let R or Python tools take over the statistical tasks.
Subio Platform and bioinformatics tools written in R or Python complement each other rather than compete. By combining them, you can build a comprehensive analysis system at a very reasonable cost.
See the last part of this movie.


University College Cork, Lab Researcher
It is an excellent user interface and the tutorials are brilliant.

Univ. of Copenhagen, Professor/Manager
This subio software platform is very easy to handle, even you include several hundred patients. The response is extremely fast compare with other similar softwares. Al...

RIKEN, professor/manager
Very flexible and powerful solution. Great technical support. VERY good advisory support!