GSE164898 is a data set using the 10X Genomics protocol. They say it can separately count mRNAs of thousands of cells from a sample tissue, but there are only tens of thousands of reads per cell. The dynamic range of RNA-Seq depends on the number of reads. With only tens of thousands of reads, you should expect that it can only capture the expression levels of a few dozen very highly expressed genes. Now let's take a look at the actual data.
First of all, the dynamic ranges of each cell are tremendously different, as is evident if you visualize it with Subio Platform. For example, on the left in the figure below are two samples from the group with the narrowest dynamic range, and on the right are two from the broadest dynamic range. Such difference in dynamic range is a dominant effect on the expression profiles of this data and doesn't reflect the alteration in the biological context as claimed in the paper.
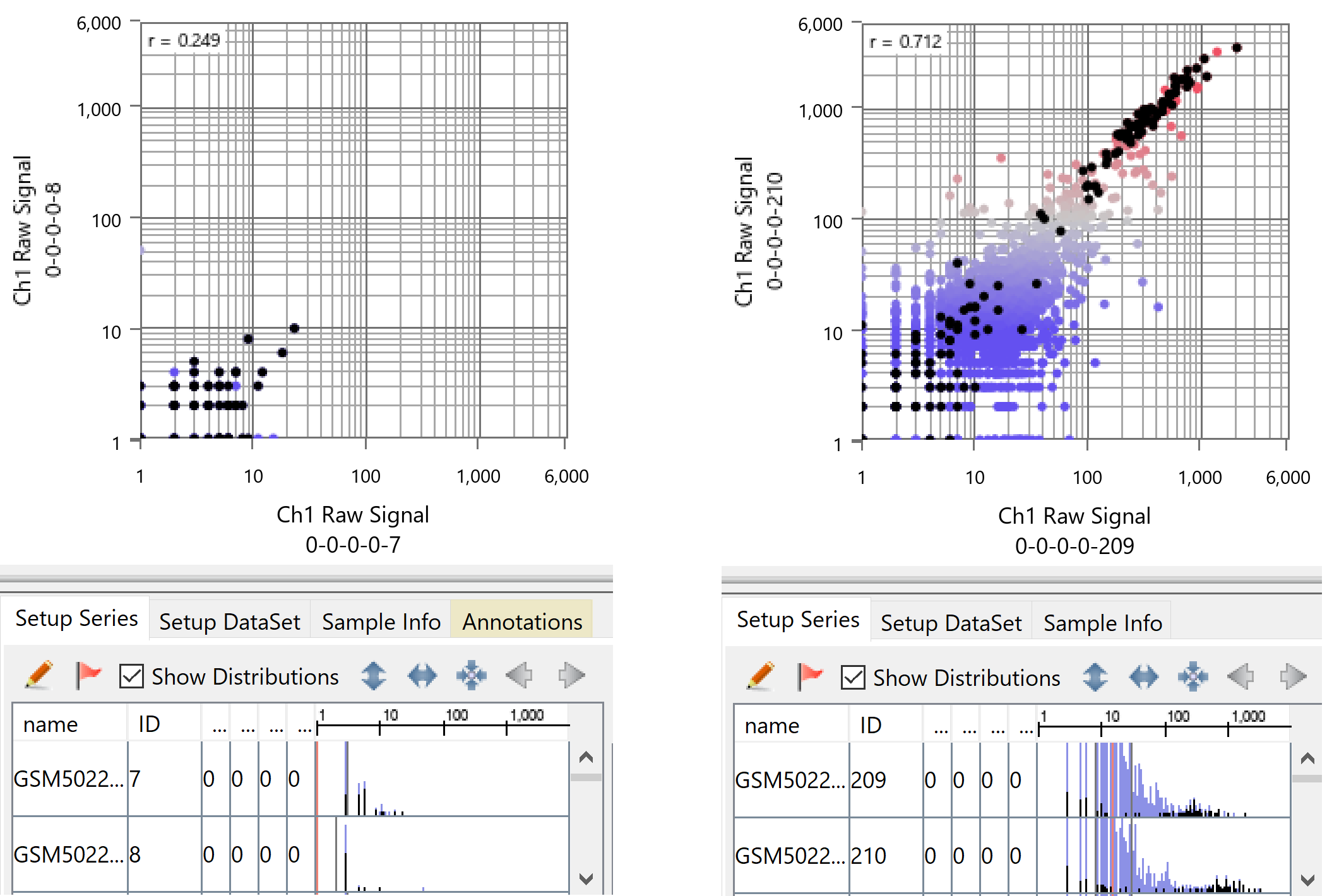
Also, you can see the border between the signal and noise ranges lays around count values of 10 to 40. In the left chart, all measurements are in the noise region. And in the right image, only about 300 genes are in the signal region. The measured count values are unreliable in the noise range, but they can also be detected or not by chance. In other words, you can't conclude that a gene doesn't express in a cell even if the count is 0. By the way, black dots represent ribosomal proteins. A gene with a count value of 1 or more suggests it is as super-highly expressing as ribosomal proteins.
Experimental biologists tend to misunderstand the images generated by Loupe Browser software as something like immunostained FISH images. However, it shows only super-highly-expressed genes captured by chance. It is essential to understand that this is utterly different from high-sensitive FISH because there is still a good chance that even genes that do not appear to be highly expressing.
Single-cell RNA-Seq is still technically limited and is not mature as regular RNA-Seq. Thus, Subio recommends that you assess whether the protocol you are adopting is capable of your research objectives before deciding. Please use our Data Analysis Service for assessments.