This is a preparation guide for using the FASTQ to VCF tool.
Install GATK4
Please finish setting up the pipeline for RNA-Seq FASTQ file processing on macOS first, because this pipeline also uses the same tools of fastp and HISAT2. What you add is only one more software, GATK4, using bioconda. So, just run the following command with the terminal to do it.
$ conda install -c bioconda gatk4
The Environment Set-up Panel
Setting up the pipeline is like doing it for the RNA-Seq FASTQ file processing. Settings for the fastp and HISAT2 executable are the same. The GATK Resource Bundle is available only for human (b37/hg19 and Grch38/hg38) but not for other organisms.
However, the indexes provided by the HISAT2 website cause the error on GATK execution because their chromosome names are like "1", not "chr1." So, download the "chr" added version of GRCh38 HISAT2 indexes and GTF file that we added "chr" to chromosome names. Please uncompress the zip files before you use them.
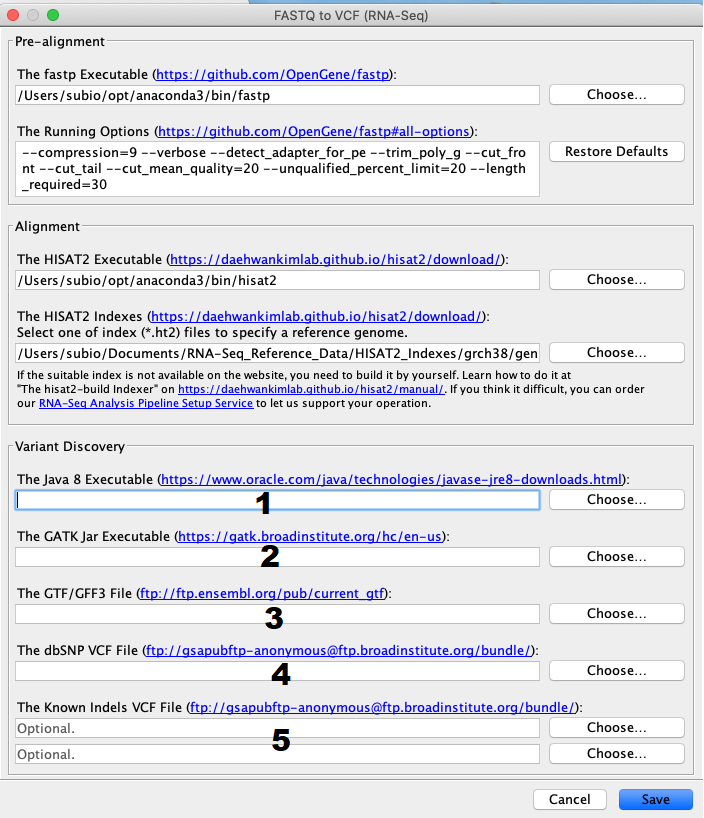
1. Java 8 Executable
Go to the anaconda3 install directory, and select /bin/java
2. GATK jar Executable
Go to the anaconda3 install directory, and select pkgs/gatk[version]/share/gatk[version]/gatk-package[version]-local.jar
3. GTF/GFF3 File
You can download the VCF file of GRCh38 version from here. This GTF file is modified version of the original provided by EBI. Chromosome names have "chr." And to make this work, you need to use it together with the "chr" added version of HISAT2 index.
4. dbSNP VCF File
Get it from gcp-public-data--broad-references on Google Cloud. For GRCh38 data, please download Homo_sapiens_assembly38.dbsnp.vcf under the hg38/v0 folder.
5. Known Indels VCF File
You can get them from the same directory as step4. Download Homo_sapiens_assembly38.known_indels.vcf and Mills_and_1000G_gold_standard.indels.hg38.vcf under the hg38/v0 folder. Please note that you have to go to the second page to see them.
Troubleshooting of the files of Step 4 to 5.
Though the names of those files end with ".vcf.gz" on Goole Cloud, downloaded files might look like ".vcf" or something else. Both raw vcf and gz compressed files are accepted, but you may need to correct extensions.
A workaround for the error "env: python: No such file or directory" on recent macOS.
On executing hisat2-inspect, it halts because the error says it can't find Python. If you see such an error, please close the Subio Platform software. And then open it with the following command from Terminal.
open /Applications/Subio/Subio.app
If you want to analyze on hg19
If you analyze on hg19, please download the original HISAT2 Index (grch37_genome.tar.gz) and GRCh37 GTF file. And use the following dbSNP and Indels files.