Firstly, The Tree View makes everything slow. The Line Graph View can also slow down as the number of samples grows. So, to keep showing Scatter Plot (Measurements) View can significantly improve the performance.
When you handle a large data set, the software inevitably slows down. For better performance, the large memory (RAM), high speed SSD (PCI Express 4.0 x4), and fast CPU is the solution. And please make sure that you have enough free disk space on the C drive .
If you don't have such options, please try the followings.
Close Other Software:
If the computer runs much software, try to close them as much as possible. If many software runs, the Subio Platform can't get enough memory space.
Disable Views and Tabs:
You can disable views and tabs from the View menu to omit drawing them.
Turning Views and Tabs On/Off
Subio Platform slows down if you load a very large data sets like million measurements or thousands of samples.
You can improve responsiveness by turning Views and tabs off. Subio Platform works like operation console without viewers.
After you complete anaylsis taks, you can turn viewrs and tabs on.
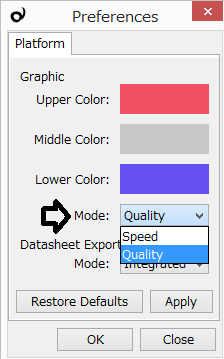
Drawing Mode:
Preferences window is accessible from the Platform or Plug-in menu. The "Quality" mode draws smooth graphics, and it costs high for the performance. So, you can switch to the "Speed" mode to have Subio Platform quickly draw rough graphics.
Fixing Scales of Chart:
The default setting is "max" and "min," which make Subio Platform search max and min number, and it consumes resources. You can set the actual number for max and min to avoid it.