This is a guide for preparing for importing RNA-Seq FASTQ files to Subio Platform on a Mac computer. If you use a Windows10 machine, please go to the guide for Windows10.
Subio Platform utilizes the following tools to process the RNA-Seq FASTQ files.
Instruction:
- Download and install the Anaconda . Even for M1 processor machines, you must explicitly download a regular version, which is NOT marked as "(M1)" because many bioinformatics tools still don't support the M1 environment. If you already have the M1 version of Anaconda, please uninstall it.
- Launch the Terminal and run the command to install fastp, HISAT2, and StringTie. Please notice that you need the option to specify version 0.22.0 for the fastp installation command.
$ conda install -c bioconda fastp==0.22.0 $ conda install -c bioconda hisat2 $ conda install -c bioconda stringtie
- Run the command to show the path of the executable files of fastp, HISAT2 and StringTie.
$ which fastp $ which hisat2 $ which stringtie
- Copy & paste the path of the executable files in the Subio Platform's setting panel.
- Download the HISAT2 indexes of the suitable organism.
- Download the GTF file of the target organism, of the same genome version as the HISAT indexes.
- Set the path to the indexes and GTF file on the Subio Platform's setting panel.
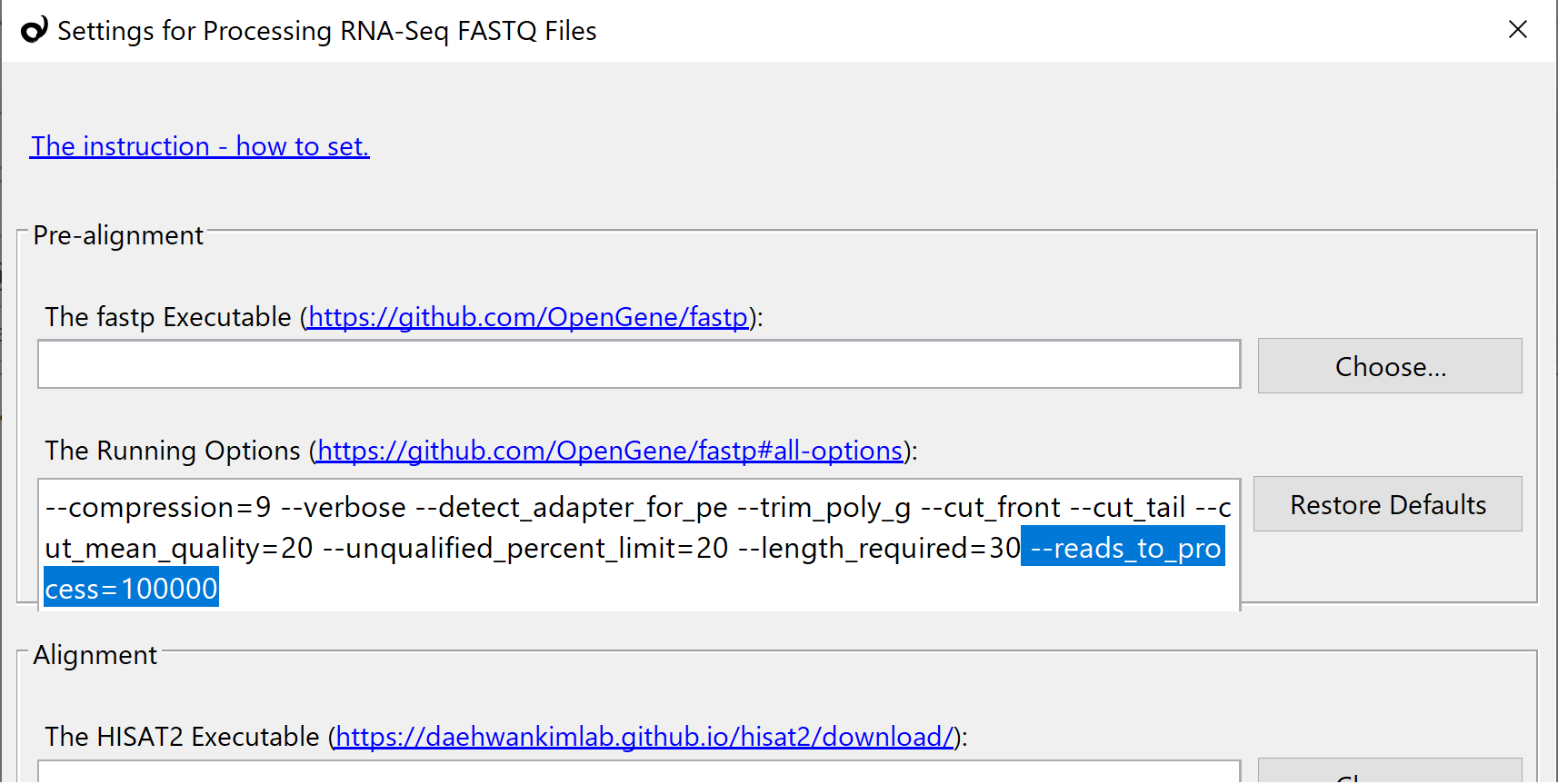
The FASTQ file processing takes a long time. So add the following option in the fastp setting. It limits the number of read-sequences to process, and it tells you the pipeline works fine or not within a couple of minutes. After the confirmation, delete all imported samples. And then, delete this option to execute on the whole.
--reads_to_process=100000
We confirmed that the pipeline works correctly with the following versions. The different versions of the tools might cause error.
- fastp 0.22.0
- HISAT2 2.1.0
- StringTie 2.1.1
We notice that HISAT2 2.2.0 is a bit trickier to make it work because you have to remove all spaces from the paths to the index and GTF.
When you installed another version of fastp, and it doesn't work, please install fastp 0.22.0 with the anaconda command shown above to overwrite.